
WINTER SCHOOL ALGORITHMS
IN STRUCTURAL BIOINFORMATICS :
MODELING ANTIBODIES AND THEIR COMPLEXES
14-18 november 2016, Inria Sophia Antipolis, France

[Daily Schedule]
graphical-abstract-1vfb-iatoms-fcazals.jpg
- Morning session: lecture from 9am to noon.
- Afternoon session: practical from 2pm to 5pm.
[Monday the 14th] Standards and tools for sequences and 3D structures of immunoglobulins: IMGT®, the international ImMunoGeneTics information system
Instructors: Marie-Paule Lefranc and Patrice Duroux, CNRS and University of Montpellier -- IMGT
Slides:
Topics:
- Immunoglobulins: a primer,
- IMGT, the international ImMunoGeneTics information system,
- Analyzing sequences of IG,
- Analyzing structures of IG.
[Tuesday the 15th] Basic concepts and novel algorithms for modeling antibodies and their interactions
Instructors: Charles H. Robert, CNRS / IBPC, France Juan Cortés, LAAS / CNRS, Toulouse, France
Slides:
Topics:
- Biophysics of protein interactions: a primer,
- Docking algorithms,
- Modeling proteins as robots to enhance conformational sampling,
- Cost-based motion-planning algorithms to efficiently compute transition paths of highly flexible molecules.
[Wednesday the 16th] Modeling antibody - antigen complexes: predicting binding affinity and interaction specificity
Instructor: Frédéric Cazals , Inria / Algorithms-Biology-Structure, Inria Sophia Antipolis
Slides:
Topics:
- Modeling interfaces of macro-molecular complexes,
- Basics in regression analysis,
- Basics in geometric modeling with Voronoi diagrams,
- Predicting binding affinities for IG - Ag complexes,
- Analyzing the interaction specificity of IG,
- Corresponding tools in the Structural Bioinformatics Library.
[Thursday the 17th] In-silico structure based design of antibodies
Instructor: Charlotte Deane, Oxford University
Slides:
Topics:
- SABDAB – the antibody structure database,
- ABodybuilder -- automated antibody structure prediction with data-driven accuracy estimation,
- Orientating the VH and VL domains,
- Predicting CDR-H3 rapidly and accurately,
- Paratopes and Epitopes – predicting and designing binding.
[Friday the 18th] Visualizing binding assay data using antigenic cartography
Instructor: David F. Burke, Cambridge University
Slides:
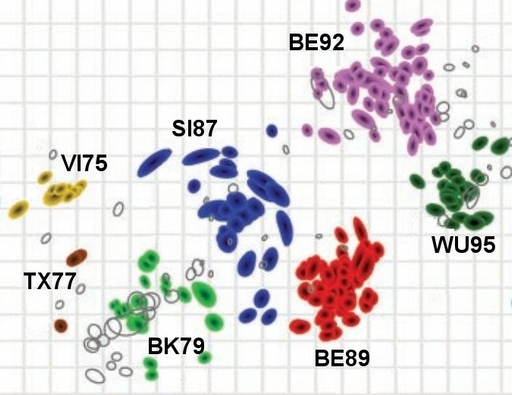
Topics:
- Theory of antigenic cartography,
- Influenza viruses,
- Example application: understanding the antigenic drift of influenza viruses.
| Sponsors |  |
 |
 |
 |
| webmaster |